Examples¶
First example: extract a time-series using the API and plot it with matplotlib¶
from geextract import ts_extract, get_date
from datetime import datetime
import numpy as np
import matplotlib.pyplot as plt
plt.figure(figsize=(10,5))
# Extract a Landsat 7 time-series for a 500m radius circular buffer around
# a location in Yucatan
lon = -89.8107197
lat = 20.4159611
raw_dict = ts_extract(lon=lon, lat=lat, sensor='LE7',
start=datetime(1999, 1, 1), radius=500)
# Function to compute ndvi from a dictionary of the list of dictionaries returned
# by ts_extract
def ndvi(x):
try:
return (x['B4'] - x['B3']) / (x['B4'] + x['B3'])
except:
pass
# Build x and y arrays and remove missing values
x = np.array([get_date(d['id']) for d in raw_dict])
y = np.array([ndvi(d) for d in raw_dict], dtype=np.float)
x = x[~np.isnan(y)]
y = y[~np.isnan(y)]
# Make plot
plt.plot_date(x, y, "--")
plt.plot_date(x, y)
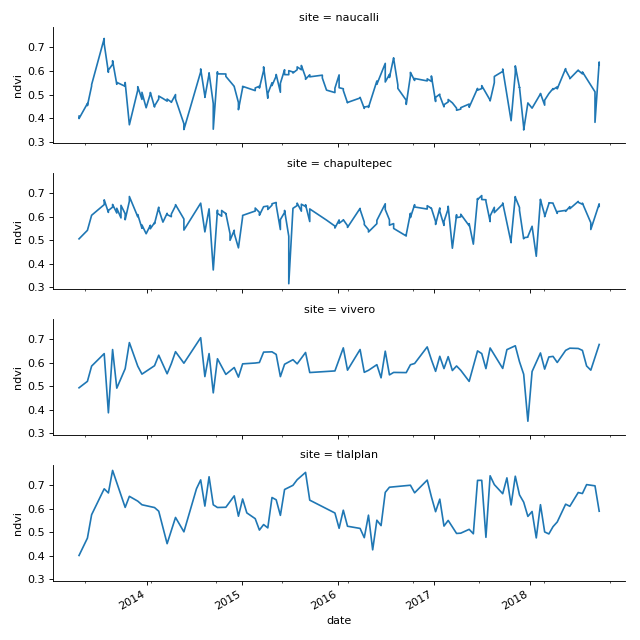
plt.title("Landsat 7 NDVI time-series Uxmal")
plt.ylabel("NDVI (-)")
plt.grid(True)
plt.show()
(Source code, png, hires.png, pdf)
Second example: extract a time-series using the command line and read it in R¶
gee_extract.py -s LT5 -b 1980-01-01 -lon -89.8107 -lat 20.4159 -r 500 -db /tmp/gee_db.sqlite -site uxmal -table col_1
gee_extract.py -s LE7 -b 1980-01-01 -lon -89.8107 -lat 20.4159 -r 500 -db /tmp/gee_db.sqlite -site uxmal -table col_1
gee_extract.py -s LC8 -b 1980-01-01 -lon -89.8107 -lat 20.4159 -r 500 -db /tmp/gee_db.sqlite -site uxmal -table col_1
Running the three above commands gives you the following terminal output. Records refer to individual time steps at which Landsat observations were extracted. Note that some records may be “empty” due to absence of useful data after the cloud masking step. The amount of useful data of the entire time-series is therefore likely to be less than the reported extracted records.
Extracted 148 records from Google Eath Engine
Extracted 231 records from Google Eath Engine
Extracted 82 records from Google Eath Engine
A sqlite database has been created at /tmp/gee_db.sqlite; it contains a table named col_1 (for “collection 1”) that can be read as an R dataframe using tools like dplyr.
library(dplyr)
library(DBI)
# Open database connection (requires dbplyr and RSQLite packages, DBI installed via dbplyr)
con <- dbConnect(RSQLite::SQLite(), dbname = "/tmp/gee_db.sqlite")
dbListTables(con)
df <- tbl(con, 'col_1') %>% collect()
df
In that case the table contains only one time-series (uxmal site); we are therefore loading the whole table in memory using collect() without additional filtering query.
[1] "col_1"
# A tibble: 390 x 11
index blue green id nir red swir1 swir2 time sensor site
<int> <dbl> <dbl> <chr> <dbl> <dbl> <dbl> <dbl> <chr> <chr> <chr>
1 0 434 635 LT05_020046_19850206 2077 675 2267 1281 1985-02-06 LT5 uxmal
2 1 370 664 LT05_020046_19850427 2883 588 2136 1128 1985-04-27 LT5 uxmal
3 2 385 592 LT05_020046_19860108 2732 553 2010 953 1986-01-08 LT5 uxmal
4 3 555 748 LT05_020046_19860313 1971 823 2497 1479 1986-03-13 LT5 uxmal
5 4 574 804 LT05_020046_19860414 2216 919 2751 1701 1986-04-14 LT5 uxmal
6 5 790 1084 LT05_020046_19860703 3852 955 2205 1121 1986-07-03 LT5 uxmal
7 6 546 858 LT05_020046_19860820 3876 730 1968 896 1986-08-20 LT5 uxmal
8 7 334 560 LT05_020046_19861007 2694 532 2088 1072 1986-10-07 LT5 uxmal
9 8 321 539 LT05_020046_19861023 2550 524 2064 1082 1986-10-23 LT5 uxmal
10 9 590 832 LT05_020046_19870417 2390 891 2752 1660 1987-04-17 LT5 uxmal
# ... with 380 more rows
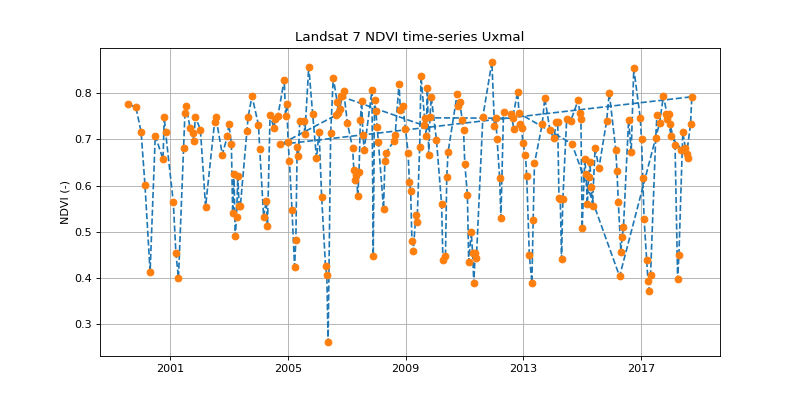
This dataframe (or tibble) can now be used as the base for all kind of data analysis in R. Here we’ll make some simple plots using the ggplot2 package.
library(ggplot2)
library(tidyr)
df %>% mutate(ndvi = (nir - red) / (nir + red)) %>%
ggplot(aes(time, ndvi)) +
geom_line() +
geom_point(aes(col = sensor)) +
theme_bw()
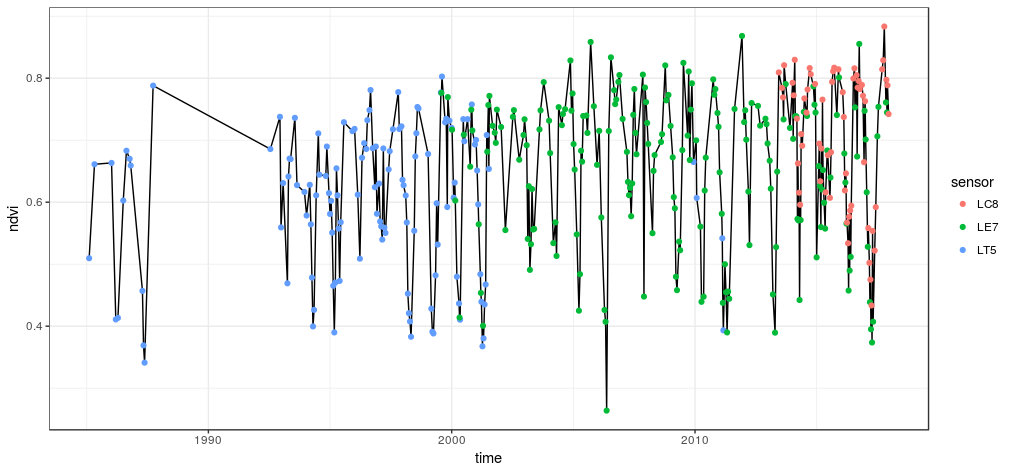
df %>% gather(key, value, -c(time, index, sensor, site, id)) %>%
ggplot(aes(time, value)) +
geom_line() +
geom_point(aes(col = sensor)) +
facet_grid(key ~ ., scales = 'free') +
theme_bw()
The idea when working with multiple sites is to append them all to the same database table and use sql (raw or via dplyr) to filter the desired data. Ordering time-series for multiple sites can be done in batch thanks to the gee_extract_batch.py command (run gee_extract_batch.py --help to see the detailed usage instructions). Here we will simply append another site to the sqlite table by re-running the gee_extract.py commands with different coordinates.
gee_extract.py -s LT5 -b 1980-01-01 -lon 4.7174 -lat 44.7814 -r 500 -db /tmp/gee_db.sqlite -site rompon -table col_1
gee_extract.py -s LE7 -b 1980-01-01 -lon 4.7174 -lat 44.7814 -r 500 -db /tmp/gee_db.sqlite -site rompon -table col_1
gee_extract.py -s LC8 -b 1980-01-01 -lon 4.7174 -lat 44.7814 -r 500 -db /tmp/gee_db.sqlite -site rompon -table col_1
Extracted 104 records from Google Eath Engine
Extracted 494 records from Google Eath Engine
Extracted 193 records from Google Eath Engine
Now the col_1 sqlite table contains time-series for two different sites (uxmal and rompon). Loading the time-series of a single site can be done thanks to the filter() dplyr verb.
df <- tbl(con, 'col_1') %>%
filter(site == 'rompon') %>%
collect() %>%
mutate(time = as.Date(time))
df
# A tibble: 513 x 11
index blue green id nir red swir1 swir2 time sensor site
<int> <dbl> <dbl> <chr> <dbl> <dbl> <dbl> <dbl> <date> <chr> <chr>
1 0 1023 1179 LT05_196029_19840409 2438 1193 2096 1329 1984-04-09 LT5 rompon
2 1 822 1035 LT05_196029_19840425 2561 987 2025 1125 1984-04-25 LT5 rompon
3 4 451 715 LT05_196029_19840612 3481 582 1893 870 1984-06-12 LT5 rompon
4 6 481 691 LT05_196029_19840815 2935 624 1799 866 1984-08-15 LT5 rompon
5 7 370 590 LT05_196029_19840831 2880 534 1736 818 1984-08-31 LT5 rompon
6 8 358 580 LT05_196029_19841002 2833 510 1560 708 1984-10-02 LT5 rompon
7 10 408 642 LT05_196029_19841119 2491 656 1693 817 1984-11-19 LT5 rompon
8 13 744 991 LT05_196029_19850327 2284 1033 2046 1239 1985-03-27 LT5 rompon
9 16 579 845 LT05_196029_19850530 3678 697 2114 999 1985-05-30 LT5 rompon
10 17 546 800 LT05_196029_19850615 3697 654 1928 933 1985-06-15 LT5 rompon
# ... with 503 more rows
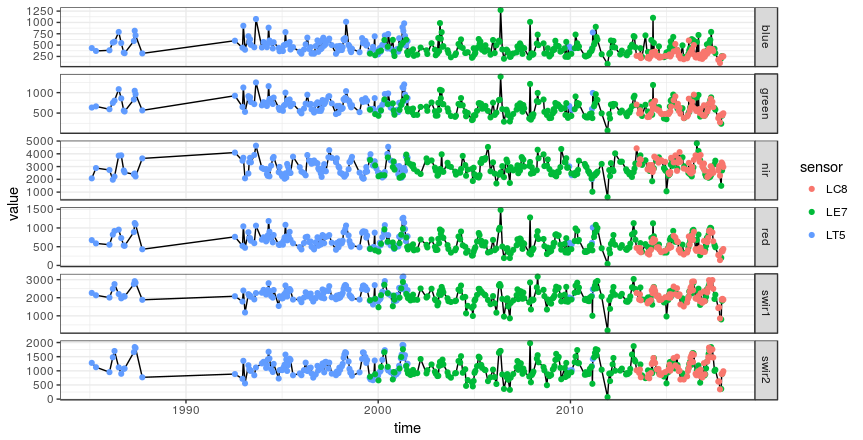
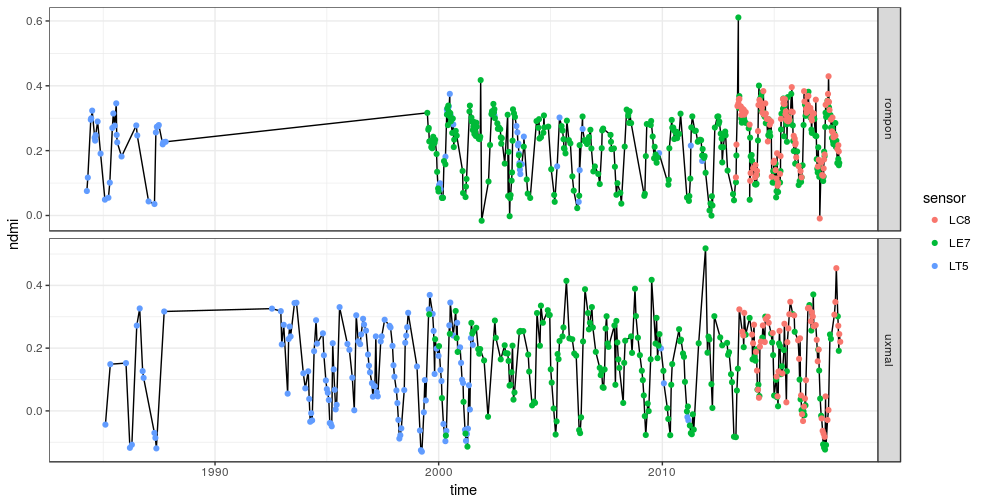
It is also possible to load the entire table to for instance plot the two time-series side by side.
df <- tbl(con, 'col_1') %>%
collect() %>%
mutate(time = as.Date(time))
df %>% mutate(ndmi = (nir - swir1) / (nir + swir1)) %>%
ggplot(aes(time, ndmi)) +
geom_line() +
geom_point(aes(col = sensor)) +
facet_grid(site ~ ., scales = 'free') +
theme_bw()
Third example: Extract time-series for each feature of a polygon feature collection¶
The file cdmx_parks.gpkg contains contours of some of the parks of Mexico City. The example below extracts the spatially aggregated time-series for each of these parks, writes the results to a sqlite database and reads the data back into python for plotting.
from geextract import ts_extract, relabel, date_append, dictlist2sqlite
import fiona
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import sqlite3
from datetime import datetime
import os
try:
os.remove('/tmp/landsat_cdmx.sqlite')
except:
pass
# Open feature collection generator
with fiona.open('data/cdmx_parks.gpkg', layer='parks') as src:
# Iterate over feature collection
for feature in src:
# Extract time-series
ts_0 = ts_extract(sensor='LC8', start=datetime(2012, 1, 1),
feature=feature)
ts_1 = relabel(ts_0, 'LC8')
ts_2 = date_append(ts_1)
# Write dictionnary list to sqlite database table
dictlist2sqlite(ts_2, site=feature['properties']['name'],
sensor='LC8', db_src='/tmp/landsat_cdmx.sqlite',
table='cdmx')
# REad the data back into a pandas dataframe
conn = sqlite3.connect("/tmp/landsat_cdmx.sqlite")
df = pd.read_sql_query('select * from cdmx', conn)
df['date'] = pd.to_datetime(df['time'], format='%Y-%m-%d')
df['ndvi'] = (df.nir - df.red) / (df.nir + df.red)
# Make facetgrid plot
g = sns.FacetGrid(df, row='site', aspect=4, size=2)
def dateplot(x, y, **kwargs):
ax = plt.gca()
data = kwargs.pop("data")
data.plot(x=x, y=y, ax=ax, grid=False, **kwargs)
g = g.map_dataframe(dateplot, "date", "ndvi")
plt.show()
(Source code, png, hires.png, pdf)